
Plot design optimization
Anika Seppelt
Source:vignettes/plot_design_optimization.Rmd
plot_design_optimization.RmdThe function metrics.variables used for the calculation
of stand-level variables and metrics (see vignette “Stand-level”)
requires arguments specifying the plot designs and sizes. If the optimal
plot design and size for the calculation of stand-level variables is not
known, the optimal plots design for the corresponding TLS data can be
determined by two different approaches implemented in FORTLS. The
approaches depend on whether field data for the sample plots is
available or not.
Estimating optimal plot size without field data
If no field data is available, the function
estimation.plot.size can be applied to
determine the optimal plot design and size. This function uses the data
frame containing the list of detected trees (introduced in
tree.tls) and estimates stand-level
density
(,
trees/ha) and basal area
(,
m/ha)
for many simulated differently-sized plots and the three plot designs
(circular fixed area, k-tree and angle-count) by increasing continuously
their sizes.
Thus, circular fixed area plots with increasing radius (increment of
0.1 m) to the maximum radius defined by
radius.max in
plot.parameters (by default set to 25, if
radius is larger than furthest tree, the horizontal distance to this
furthest tree is considered as maximum radius) will be simulated and for
each plot, the variables (N and G) are estimated. Similarly, k-tree
plots with tree numbers
()
ranging from 1 to k.max (specified in
plot.parameters, default value set to 50 or total number of
trees in the plot) and angle-count plots with increasing basal area
factor (BAF, increments of 0.1
m/ha)
to the maximum value specified by BAF.max
in plot.parameters (set to 4 by default) are simulated and
the respective stand-level variables are calculated. Optionally, the
minimum diameter at breast height
(dbh.min, in cm) to include the trees in
the estimations can be defined. By default the minimum
is set to 4 cm.
The function generates size-estimation charts i.e., plots showing the
estimated stand-level density
()
and basal area
()
on the
axes respective to the different plot sizes
(
axes). The estimations will be performed for simulated plots
corresponding to all sample plots. By default the output graphs will
contain one line for each sample plot. When
average is set to TRUE, the
average of all estimations (for all plots) as a continuous line and the
standard deviation as grey shaded area will be drawn instead of multiple
lines for each sample plot. One chart for each plot design is drawn by
default. If all.plot.designs is set to
TRUE, the line charts of all three plot design will be
drawn in one graph with different colours for each plot design.
estimation.plot.size(tree.tls = tree.tls,
plot.parameters = data.frame(radius.max = 25, k.max = 50, BAF.max = 4),
dbh.min = 4,
average = TRUE, all.plot.designs = FALSE)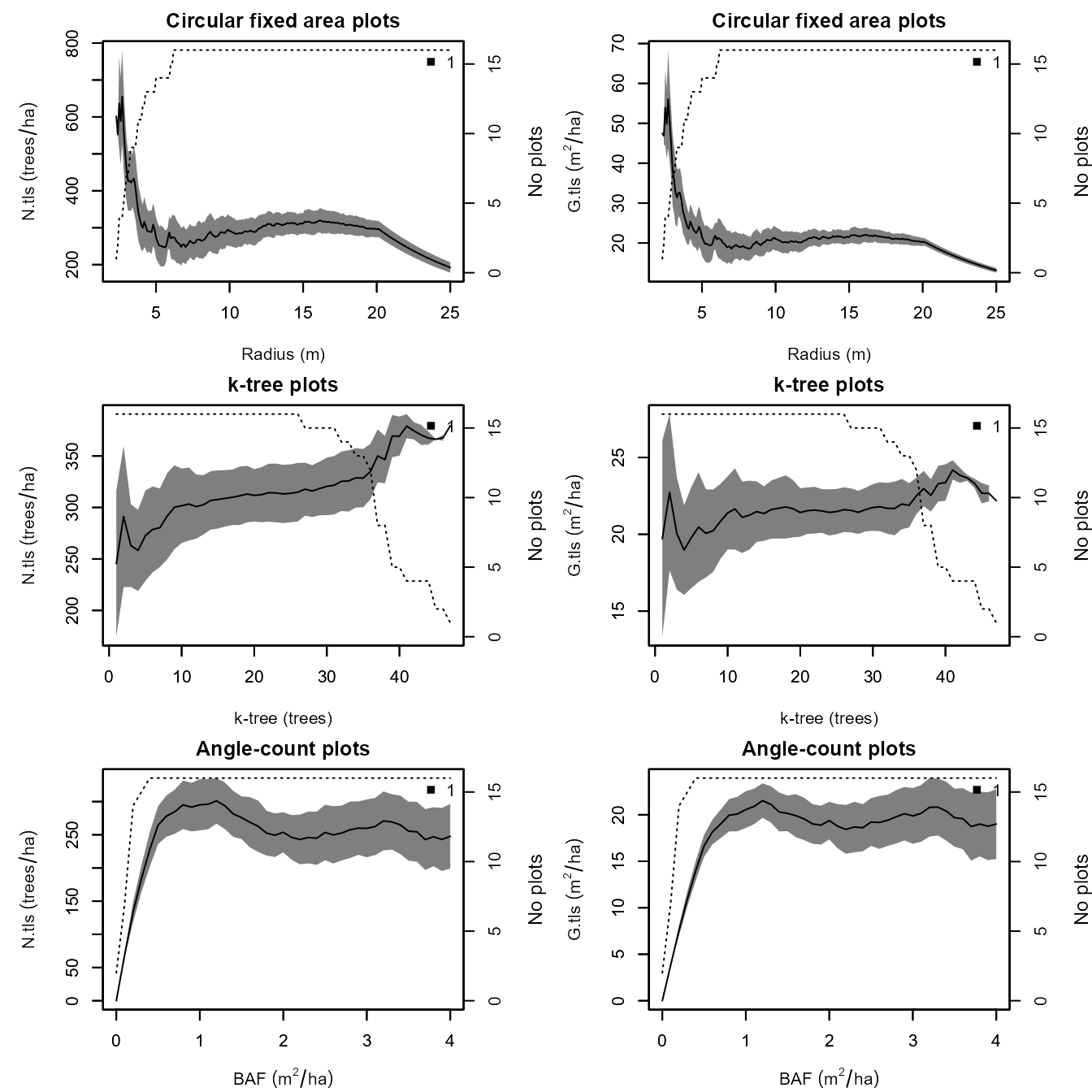
The continuous line represents the average over all sample plots (i.e. 16 plots in the example shown here) of the estimated density () on the left and the basal area () on the right. The dotted line indicates the number of sample plots. This figure helps to find suitable plot designs for the calculation of stand-level metrics and variables. The optimal plot design and size should be chosen within a range where the estimated values for and reach a stable level. A too small plot leads to high errors of estimation, since only few trees enter the plot and therefore the sample is too small. In the example above, the basal area estimated for fixed area plots with radius smaller than 5 m is much higher (around 40-50 m/ha) than the true value (around 20 m/ha). On the other hand, too large plots come along with systematic errors due to occlusion of trees. Therefore, the basal area in the same example of fixed area plots with radius bigger than 20 m is estimated lower than the true value. In order to avoid both types of errors, the figure helps to find a plot size range with stable values.
Validation with field data and optimizing plot design
When data from field measurements are available for the same sample
plots, the TLS-based estimates can be validated and the optimal plot
designs can be found applying functions implemented in FORTLS. In the
first step of the optimization process, the function
simulations simulates plots with
incremental size and computes the corresponding stand-level metrics and
variables (similar to the function metrics.variables, see
“Stand-level” vignette). Based on the simulated data, two different
processes can be performed. First, the bias between TLS data and field
data for each individual estimated variable can be assessed with the
function relative.bias. Second,
correlations between all estimated variables and metrics based on
TLS-data (output data of the simulations function) and the
variables estimated from field data can be calculated with the
correlations function. This function
calculates both the Pearson and Spearman correlation coefficients. To
visualize the correlation coefficients, heat maps can be drawn with the
optimize.plot.design function.
Plot simultaion and estimation of metrics and variables
The simulations function is applied as
follows:
simulations <- simulations(tree.tls = tree.tls, tree.ds = tree.ds, tree.field = tree.field,
plot.design = c("fixed.area", "k.tree", "angle.count"),
plot.parameters = data.frame(radius.max = 25, k.max = 50, BAF.max = 4),
scan.approach = "single", var.metr = list(tls = NULL, field = NULL),
dbh.min = 4, h.min = 1.3, max.dist = Inf,
dir.data = dir.data, save.result = FALSE, dir.result = NULL)The input data frames
Both TLS and field data from the same sample plots are required to
compute the function. The TLS data introduced in
tree.tls should have the same format as
the data frame returned from the tree.detection.single.scan
and tree.detection.multi.scan functions. Thus, each row
must correspond to a detected tree and it must contain at least the
following columns: id, file,
tree, x, y,
phi.left, phi.right,
horizontal.distance, dbh,
num.points, num.points.hom,
num.points.est, num.points.hom.est and
partial.occlusion. The data frame containing the field data
must be inserted in the argument
tree.field. Similar to the TLS data table,
each row must correspond to a tree (specified in the columns
id and tree) and the values for
horizontal.distance, dbh,
total.height and an integer value indicating whether the
tree is dead (1) or alive (NA, specified in dead) must be
included in the data frame.
When the distance sampling method for correction of occlusion effects
was applied (function distance.sample, see “Stand-level”
vignette), a list with the results in the output data frames from the
aforesaid function can be introduced in
tree.ds. The list must contain at least
the data frame tree with the detection probabilities
(P.hn, P.hn.cov, P.hr and
P.hr.cov) for each tree. By default tree.ds is
set to NULL and as a result, the calculations of the
variables based on occlusion correction will not be performed.
Specifying designs of simulated plots
A vector containing the names of the plot designs can specify the
plot designs ("fixed.area", "k.tree",
"angle.count") that are to be considered for the
simulations in plot.design. By default,
this argument is set to NULL and all three plots designs
will be considered.
Furthermore the argument
plot.parameters allows for manually
specifying the design of the simulated plots. Many differently-sized
plots of the plot designs specified in plot.design are
simulated. The list introduced in plot.parameters can
include the following elements to customize the generated plots. The
elements radius.max, k.tree.max and
BAF.max define the maximum radius (in m), the maximum
number of trees and the maximum BAF (in
m/ha)
respectively to which the sizes of circular fixed area, k-tree and
angle-count plots respectively should increase. By default the values
are set to radius.max = 25, k.tree.max = 50
and BAF.max = 4. The increment by which the sizes of
circular fixed area and angle-count plots sequentially increase can also
be customized by specifying the elements radius.increment
and BAF.increment respectively. The default settings are
radius.increment = 0.1 (in m) and
BAF.increment = 0.1 (in
m/ha).
An additional element of the list can be num.trees defining
the number of dominant trees per hectare (trees/ha). This value is
needed for the calculation of dominant diamters and heights and is set
to 100 trees/ha by default.
Further adjustable arguments
Similar to the other functions, the scan approach can be specified in
scan.approach and is by default set to
"multi". Metrics and variables of interest can be defined
as a vector in var.metr. Thus, only those
metrics and variables named in the vector are calculated. If not
specified, var.metr is set to NULL and all
possible metrics and variables are computed. The arguments
dbh.min,
h.min and
max.dist can optionally define the minimum
,
height and maximum distance of a tree to be included in the
calculations. The default values are dbh.min = 4 (in cm),
h.min = 1.3 (in m) and max.dist = NULL (no
maximal distance is considered).
The argument dir.data should specify
the working directory of the .txt files with the normalized reduced
point clouds (output of normalize function). If not
specified, it is set to NULL and the current working
directory is assigned to it. The output files will be saved by default,
since the argument save.result is set to
TRUE, to the directory path indicated in
dir.result. For each plot design (circular
fixed area, k-tree and angle-count plots), a .csv file is created using
the write.csv function from the utils package.
Output of the simulations function
The simulations function generates a list with one
element for each plot design. The elements are data frames containing
the simulated plot. Each row represents a simulated plot defined by
their respective plot identification number id and their
size determined by either radius, k or
BAF depending on the plot design. The columns
N, G, V, V.user,
W.user, d, dg,
dgeom, dharm, h, hg,
hgeom, hharm, d.0,
dg.0, dgeom.0, dharm.0,
h.0, hg.0, hgeom.0 and
hharm.0 display the stand-level variables based on the
field data. The remaining columns contain the stand-level variables and
metrics estimated for each plot based on the TLS data. As an example the
data frame for circular fixed area plots is shown below.
head(simulations$fixed.area)| id | radius | N | G | V | V.user | W.user | d | dg | dgeom | dharm | h | hg | hgeom | hharm | d.0 | dg.0 | dgeom.0 | dharm.0 | h.0 | hg.0 | hgeom.0 | hharm.0 | N.tls | N.hn | N.hr | N.hn.cov | N.hr.cov | N.sh | G.tls | G.hn | G.hr | G.hn.cov | G.hr.cov | G.sh | V.tls | V.hn | V.hr | V.hn.cov | V.hr.cov | V.sh | d.tls | dg.tls | dgeom.tls | dharm.tls | h.tls | hg.tls | hgeom.tls | hharm.tls | d.0.tls | dg.0.tls | dgeom.0.tls | dharm.0.tls | h.0.tls | hg.0.tls | hgeom.0.tls | hharm.0.tls | n.pts | n.pts.est | n.pts.red | n.pts.red.est | P01 | P05 | P10 | P20 | P25 | P30 | P40 | P50 | P60 | P70 | P75 | P80 | P90 | P95 | P99 | mean.z | mean.q.z | mean.g.z | mean.h.z | median.z | mode.z | max.z | min.z | var.z | sd.z | CV.z | D.z | ID.z | kurtosis.z | skewness.z | p.a.mean.z | p.a.mode.z | p.a.2m.z | p.b.mean.z | p.b.mode.z | p.b.2m.z | CRR.z | L2.z | L3.z | L4.z | L3.mu.z | L4.mu.z | L.CV.z | median.a.d.z | mode.a.d.z | weibull_c.z | weibull_b.z | mean.rho | mean.q.rho | mean.g.rho | mean.h.rho | median.rho | mode.rho | max.rho | min.rho | var.rho | sd.rho | CV.rho | D.rho | ID.rho | kurtosis.rho | skewness.rho | p.a.mean.rho | p.a.mode.rho | p.b.mean.rho | p.b.mode.rho | CRR.rho | L2.rho | L3.rho | L4.rho | L3.mu.rho | L4.mu.rho | L.CV.rho | median.a.d.rho | mode.a.d.rho | weibull_c.rho | weibull_b.rho | mean.r | mean.q.r | mean.g.r | mean.h.r | median.r | mode.r | max.r | min.r | var.r | sd.r | CV.r | D.r | ID.r | kurtosis.r | skewness.r | p.a.mean.r | p.a.mode.r | p.b.mean.r | p.b.mode.r | CRR.r | L2.r | L3.r | L4.r | L3.mu.r | L4.mu.r | L.CV.r | median.a.d.r | mode.a.d.r | weibull_c.r | weibull_b.r |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 2.5 | 509.2958 | 53.87560 | 485.1621 | 385.4172 | 175.3602 | 36.7 | 36.7 | 36.7 | 36.7 | 16.6 | 16.6 | 16.6 | 16.6 | 36.7 | 36.7 | 36.7 | 36.7 | 16.6 | 16.6 | 16.6 | 16.6 | 509.2958 | 535.4928 | 535.6553 | 553.4044 | 547.1048 | 509.2958 | 51.99859 | 54.67328 | 54.68987 | 56.50203 | 55.85885 | 51.99859 | 383.8610 | 403.6059 | 403.7284 | 417.1060 | 412.3579 | 383.8610 | 36.05502 | 36.05502 | 36.05502 | 36.05502 | 13.32373 | 13.32373 | 13.32373 | 13.32373 | 36.05502 | 36.05502 | 36.05502 | 36.05502 | 13.32373 | 13.32373 | 13.32373 | 13.32373 | 4752.000 | 324.3204 | 28.33333 | 31.89828 | 1.06937 | 8.61100 | 9.339 | 9.590 | 9.694 | 9.824 | 10.110 | 10.321 | 11.033 | 12.6206 | 13.646 | 13.767 | 14.188 | 15.06200 | 15.238 | 11.00676 | 11.34071 | 10.354835 | 7.979817 | 10.321 | 10.102 | 25.396 | 0.102 | 7.463079 | 2.731864 | 0.2481988 | 25.294 | 3.952 | 6.099774 | -1.117082 | 40.05622 | 60.49542 | 98.11661 | 59.94378 | 39.41439 | 1.8818861 | 0.4334052 | 8556406 | 103592905 | 1293244862 | -178939297 | 2951898465 | 1.3e-06 | 1.4267575 | 0.9047575 | 4.578460 | 12.04913 | 1.647650 | 1.719348 | 1.571360 | 1.493911 | 1.586860 | 0.5915731 | 2.499989 | 0.5915731 | 0.2414102 | 0.4913351 | 0.2982035 | 1.908416 | 0.8601007 | 1.810289 | 0.1171962 | 46.19790 | 99.99850 | 53.80210 | 0 | 0.6590631 | 196670.2 | 377892.9 | 765026.4 | -594229.5 | 1477928 | 8.4e-06 | 0.4246912 | 1.056077 | NA | NA | 11.18274 | 11.47030 | 10.77472 | 10.101851 | 10.47458 | 9.178085 | 25.47709 | 2.1692727 | 6.514317 | 2.552316 | 0.2282371 | 23.30782 | 3.878540 | 5.003789 | -0.8179068 | 40.12085 | 94.35134 | 59.87915 | 5.647161 | 0.4389330 | 8753077 | 106671499 | 1339243442 | -186975775 | 3135302332 | 1.3e-06 | 1.4446079 | 2.004653 | 5.019574 | 12.17664 |
| 16 | 2.5 | 509.2958 | 44.62240 | 395.1908 | 315.3808 | 142.6170 | 33.4 | 33.4 | 33.4 | 33.4 | 16.3 | 16.3 | 16.3 | 16.3 | 33.4 | 33.4 | 33.4 | 33.4 | 16.3 | 16.3 | 16.3 | 16.3 | 509.2958 | 535.4928 | 535.6553 | 543.4899 | 539.0934 | 509.2958 | 36.97431 | 38.87619 | 38.88798 | 39.45676 | 39.13759 | 36.97431 | 284.0880 | 298.7008 | 298.7915 | 303.1616 | 300.7093 | 284.0880 | 30.40325 | 30.40325 | 30.40325 | 30.40325 | 13.93300 | 13.93300 | 13.93300 | 13.93300 | 30.40325 | 30.40325 | 30.40325 | 30.40325 | 13.93300 | 13.93300 | 13.93300 | 13.93300 | 6507.667 | 708.8804 | 89.33333 | 50.63408 | 1.98046 | 8.69900 | 9.048 | 10.592 | 10.936 | 11.038 | 11.325 | 11.533 | 11.885 | 12.1700 | 12.368 | 12.459 | 13.043 | 13.39300 | 14.568 | 11.24222 | 11.42735 | 10.862746 | 9.367235 | 11.533 | 12.445 | 14.948 | 0.102 | 4.196767 | 2.048601 | 0.1822239 | 14.846 | 1.432 | 11.224370 | -2.336350 | 63.75997 | 20.81752 | 98.98918 | 36.24003 | 79.10180 | 1.0108184 | 0.7520887 | 22982316 | 271444389 | 3247255172 | -503669713 | 8468728684 | 5.0e-07 | 0.9057775 | 1.2027775 | 6.413071 | 12.07408 | 1.666723 | 1.754167 | 1.564441 | 1.451082 | 1.709399 | 0.4703062 | 2.499988 | 0.4703062 | 0.2991372 | 0.5469344 | 0.3281495 | 2.029682 | 0.9615330 | 1.808112 | -0.1776052 | 51.89550 | 99.99943 | 48.10450 | 0 | 0.6666925 | 541557.5 | 1073007.2 | 2230061.9 | -1634862.8 | 4102985 | 3.1e-06 | 0.4802624 | 1.196417 | NA | NA | 11.40354 | 11.56120 | 11.15961 | 10.723566 | 11.66687 | 12.453883 | 14.99513 | 0.9449329 | 3.620746 | 1.902826 | 0.1668628 | 14.05020 | 1.431501 | 9.581083 | -2.0602270 | 63.49917 | 24.14316 | 36.50083 | 75.856269 | 0.7604825 | 23523874 | 280290716 | 3381548744 | -524472512 | 8950638614 | 5.0e-07 | 0.8801652 | 1.050345 | 7.052386 | 12.18588 |
| 4 | 2.6 | 470.8726 | 49.81102 | 448.5596 | 356.3398 | 162.1304 | 36.7 | 36.7 | 36.7 | 36.7 | 16.6 | 16.6 | 16.6 | 16.6 | 36.7 | 36.7 | 36.7 | 36.7 | 16.6 | 16.6 | 16.6 | 16.6 | 470.8726 | 495.0932 | 495.2434 | 511.6534 | 505.8291 | 470.8726 | 48.07562 | 50.54852 | 50.56386 | 52.23930 | 51.64464 | 48.07562 | 354.9010 | 373.1563 | 373.2696 | 385.6379 | 381.2481 | 354.9010 | 36.05502 | 36.05502 | 36.05502 | 36.05502 | 13.32373 | 13.32373 | 13.32373 | 13.32373 | 36.05502 | 36.05502 | 36.05502 | 36.05502 | 13.32373 | 13.32373 | 13.32373 | 13.32373 | 4752.000 | 324.3204 | 28.33333 | 31.89828 | 1.00941 | 3.52310 | 9.098 | 9.508 | 9.623 | 9.738 | 10.061 | 10.271 | 10.691 | 12.0940 | 13.240 | 13.727 | 14.125 | 15.04800 | 15.227 | 10.72964 | 11.13278 | 9.954041 | 7.537012 | 10.271 | 10.102 | 25.396 | 0.102 | 8.813723 | 2.968791 | 0.2766907 | 25.294 | 3.617 | 5.158607 | -1.100687 | 39.57380 | 58.06012 | 97.61027 | 60.42620 | 41.85642 | 2.3869469 | 0.4224933 | 8910081 | 107128487 | 1330448258 | -179674917 | 2887255543 | 1.2e-06 | 1.2446393 | 0.6276393 | 4.061632 | 11.82730 | 1.715051 | 1.794777 | 1.629195 | 1.541556 | 1.651641 | 0.5915731 | 2.599997 | 0.5915731 | 0.2798282 | 0.5289879 | 0.3084387 | 2.008424 | 0.9503215 | 1.749552 | 0.0754967 | 46.90851 | 99.99861 | 53.09149 | 0 | 0.6596356 | 231576.9 | 466972.4 | 992378.5 | -724516.0 | 1875788 | 7.4e-06 | 0.4713072 | 1.123477 | NA | NA | 10.93564 | 11.27652 | 10.44544 | 9.669257 | 10.41573 | 9.178085 | 25.47709 | 2.1692727 | 7.571908 | 2.751710 | 0.2516278 | 23.30782 | 3.648276 | 4.401105 | -0.8483287 | 39.43887 | 90.90429 | 60.56113 | 9.094323 | 0.4292341 | 9141658 | 110604689 | 1381277002 | -189302292 | 3102508288 | 1.2e-06 | 1.2825717 | 1.757553 | 4.509889 | 11.98177 |
| 16 | 2.6 | 470.8726 | 41.25592 | 365.3762 | 291.5873 | 131.8575 | 33.4 | 33.4 | 33.4 | 33.4 | 16.3 | 16.3 | 16.3 | 16.3 | 33.4 | 33.4 | 33.4 | 33.4 | 16.3 | 16.3 | 16.3 | 16.3 | 470.8726 | 495.0932 | 495.2434 | 502.4869 | 498.4222 | 470.8726 | 34.18483 | 35.94322 | 35.95413 | 36.47999 | 36.18490 | 34.18483 | 262.6553 | 276.1657 | 276.2495 | 280.2900 | 278.0226 | 262.6553 | 30.40325 | 30.40325 | 30.40325 | 30.40325 | 13.93300 | 13.93300 | 13.93300 | 13.93300 | 30.40325 | 30.40325 | 30.40325 | 30.40325 | 13.93300 | 13.93300 | 13.93300 | 13.93300 | 6507.667 | 708.8804 | 89.33333 | 50.63408 | 2.02500 | 8.58600 | 9.038 | 10.315 | 10.918 | 11.024 | 11.318 | 11.525 | 11.869 | 12.1570 | 12.358 | 12.456 | 13.037 | 13.39200 | 14.560 | 11.22232 | 11.40733 | 10.846357 | 9.363614 | 11.525 | 12.445 | 14.948 | 0.102 | 4.186814 | 2.046171 | 0.1823305 | 14.846 | 1.440 | 10.984884 | -2.288059 | 64.27412 | 20.61485 | 99.01514 | 35.72588 | 79.30825 | 0.9848639 | 0.7507572 | 24364239 | 287347738 | 3433378621 | -532919146 | 8945155375 | 5.0e-07 | 0.9336819 | 1.2226819 | 6.409016 | 12.05311 | 1.719710 | 1.811796 | 1.610972 | 1.489595 | 1.776552 | 0.4703062 | 2.599995 | 0.4703062 | 0.3252045 | 0.5702671 | 0.3316065 | 2.129689 | 1.0203854 | 1.808793 | -0.2034102 | 52.55830 | 99.99947 | 47.44170 | 0 | 0.6614281 | 614615.3 | 1259319.9 | 2705262.3 | -1911549.9 | 4948564 | 2.8e-06 | 0.5091441 | 1.249404 | NA | NA | 11.39275 | 11.55032 | 11.15117 | 10.723123 | 11.66348 | 12.453883 | 14.99513 | 0.9449329 | 3.615002 | 1.901316 | 0.1668882 | 14.05020 | 1.442599 | 9.359456 | -2.0148956 | 63.87088 | 24.08590 | 36.12912 | 75.913563 | 0.7597633 | 24978854 | 297407285 | 3586119564 | -556323552 | 9485674908 | 5.0e-07 | 0.9038278 | 1.061130 | 7.051229 | 12.17445 |
| 4 | 2.7 | 436.6391 | 46.18964 | 415.9483 | 330.4331 | 150.3431 | 36.7 | 36.7 | 36.7 | 36.7 | 16.6 | 16.6 | 16.6 | 16.6 | 36.7 | 36.7 | 36.7 | 36.7 | 16.6 | 16.6 | 16.6 | 16.6 | 873.2782 | 918.1976 | 918.4762 | 941.7921 | 932.5193 | 872.8359 | 83.92237 | 88.23914 | 88.26591 | 90.50658 | 89.61547 | 83.87987 | 638.9710 | 671.8382 | 672.0420 | 689.1021 | 682.3173 | 638.6474 | 34.96277 | 34.97982 | 34.94570 | 34.92864 | 13.82336 | 13.83238 | 13.81432 | 13.80530 | 36.05502 | 36.05502 | 36.05502 | 36.05502 | 13.32373 | 13.32373 | 13.32373 | 13.32373 | 9720.333 | 628.9908 | 72.00000 | 61.86391 | 1.07525 | 3.14325 | 7.160 | 9.422 | 9.560 | 9.672 | 9.994 | 10.223 | 10.551 | 11.8730 | 12.787 | 13.691 | 14.071 | 15.03400 | 15.217 | 10.51156 | 10.95263 | 9.695039 | 7.393004 | 10.223 | 10.102 | 25.396 | 0.102 | 9.467294 | 3.076897 | 0.2927156 | 25.294 | 3.227 | 4.563186 | -1.013085 | 40.66258 | 55.91363 | 97.68611 | 59.33742 | 44.00773 | 2.3113116 | 0.4139060 | 9305897 | 110969906 | 1369445141 | -182486188 | 2872952321 | 1.1e-06 | 1.1575571 | 0.4095571 | 3.816339 | 11.62778 | 1.783608 | 1.870852 | 1.688341 | 1.590305 | 1.737114 | 0.5915731 | 2.699997 | 0.5915731 | 0.3188325 | 0.5646525 | 0.3165788 | 2.108424 | 1.0150070 | 1.703296 | 0.0258272 | 48.02578 | 99.99871 | 51.97422 | 0 | 0.6605962 | 271519.1 | 572872.7 | 1273189.1 | -879967.0 | 2368671 | 6.6e-06 | 0.5077340 | 1.192035 | NA | NA | 10.74166 | 11.11126 | 10.21645 | 9.416921 | 10.37449 | 9.178085 | 25.47709 | 2.1692727 | 8.076948 | 2.841997 | 0.2645771 | 23.30782 | 3.231135 | 3.988811 | -0.7922456 | 39.60941 | 87.77957 | 60.39059 | 12.219143 | 0.4216203 | 9577417 | 114927230 | 1426118305 | -193702332 | 3118482248 | 1.1e-06 | 1.2037969 | 1.563575 | 4.267396 | 11.80698 |
| 16 | 2.7 | 436.6391 | 38.25652 | 338.8125 | 270.3882 | 122.2711 | 33.4 | 33.4 | 33.4 | 33.4 | 16.3 | 16.3 | 16.3 | 16.3 | 33.4 | 33.4 | 33.4 | 33.4 | 16.3 | 16.3 | 16.3 | 16.3 | 436.6391 | 459.0988 | 459.2381 | 465.9549 | 462.1857 | 437.3046 | 31.69951 | 33.33006 | 33.34018 | 33.82781 | 33.55417 | 31.74783 | 243.5597 | 256.0878 | 256.1655 | 259.9122 | 257.8097 | 243.9309 | 30.40325 | 30.40325 | 30.40325 | 30.40325 | 13.93300 | 13.93300 | 13.93300 | 13.93300 | 30.40325 | 30.40325 | 30.40325 | 30.40325 | 13.93300 | 13.93300 | 13.93300 | 13.93300 | 6507.667 | 708.8804 | 89.33333 | 50.63408 | 2.02200 | 8.58500 | 9.041 | 10.242 | 10.913 | 11.023 | 11.315 | 11.527 | 11.867 | 12.1530 | 12.351 | 12.453 | 13.043 | 13.43685 | 14.553 | 11.22242 | 11.40653 | 10.845990 | 9.334158 | 11.527 | 12.445 | 14.948 | 0.102 | 4.166236 | 2.041136 | 0.1818801 | 14.846 | 1.438 | 11.041497 | -2.286062 | 64.30236 | 20.46210 | 99.01174 | 35.69764 | 79.46481 | 0.9882597 | 0.7507642 | 25633166 | 302258980 | 3611012454 | -560736947 | 9412526857 | 4.0e-07 | 0.9335763 | 1.2225763 | 6.426180 | 12.05149 | 1.765761 | 1.862154 | 1.651184 | 1.522645 | 1.836115 | 0.4703062 | 2.699995 | 0.4703062 | 0.3497093 | 0.5913622 | 0.3349051 | 2.229689 | 1.0605005 | 1.819670 | -0.2151981 | 52.95336 | 99.99949 | 47.04664 | 0 | 0.6539866 | 683166.0 | 1440851.1 | 3186039.3 | -2178061.5 | 5789524 | 2.6e-06 | 0.5317053 | 1.295455 | NA | NA | 11.40089 | 11.55754 | 11.16129 | 10.737206 | 11.67122 | 12.453883 | 14.99513 | 0.9449329 | 3.596266 | 1.896382 | 0.1663363 | 14.05020 | 1.444901 | 9.360365 | -2.0056493 | 63.67499 | 24.19485 | 36.32501 | 75.804642 | 0.7603062 | 26316332 | 313490162 | 3782025311 | -586596001 | 10009343558 | 4.0e-07 | 0.9060391 | 1.052990 | 7.076416 | 12.18090 |
As described above, plots with increasing sizes are estimated and the corresponding variables an metrics are calculated. The plots are ordered from the smallest to the biggest. Plots with small radius can not be simulated for all sample plots (see table above), since trees not always enter, because the plots are too small. But plots with e.g. a radius of 20 m, can be simulated for all sample plots (see end of table below).
tail(simulations$fixed.area)| id | radius | N | G | V | V.user | W.user | d | dg | dgeom | dharm | h | hg | hgeom | hharm | d.0 | dg.0 | dgeom.0 | dharm.0 | h.0 | hg.0 | hgeom.0 | hharm.0 | N.tls | N.hn | N.hr | N.hn.cov | N.hr.cov | N.sh | G.tls | G.hn | G.hr | G.hn.cov | G.hr.cov | G.sh | V.tls | V.hn | V.hr | V.hn.cov | V.hr.cov | V.sh | d.tls | dg.tls | dgeom.tls | dharm.tls | h.tls | hg.tls | hgeom.tls | hharm.tls | d.0.tls | dg.0.tls | dgeom.0.tls | dharm.0.tls | h.0.tls | hg.0.tls | hgeom.0.tls | hharm.0.tls | n.pts | n.pts.est | n.pts.red | n.pts.red.est | P01 | P05 | P10 | P20 | P25 | P30 | P40 | P50 | P60 | P70 | P75 | P80 | P90 | P95 | P99 | mean.z | mean.q.z | mean.g.z | mean.h.z | median.z | mode.z | max.z | min.z | var.z | sd.z | CV.z | D.z | ID.z | kurtosis.z | skewness.z | p.a.mean.z | p.a.mode.z | p.a.2m.z | p.b.mean.z | p.b.mode.z | p.b.2m.z | CRR.z | L2.z | L3.z | L4.z | L3.mu.z | L4.mu.z | L.CV.z | median.a.d.z | mode.a.d.z | weibull_c.z | weibull_b.z | mean.rho | mean.q.rho | mean.g.rho | mean.h.rho | median.rho | mode.rho | max.rho | min.rho | var.rho | sd.rho | CV.rho | D.rho | ID.rho | kurtosis.rho | skewness.rho | p.a.mean.rho | p.a.mode.rho | p.b.mean.rho | p.b.mode.rho | CRR.rho | L2.rho | L3.rho | L4.rho | L3.mu.rho | L4.mu.rho | L.CV.rho | median.a.d.rho | mode.a.d.rho | weibull_c.rho | weibull_b.rho | mean.r | mean.q.r | mean.g.r | mean.h.r | median.r | mode.r | max.r | min.r | var.r | sd.r | CV.r | D.r | ID.r | kurtosis.r | skewness.r | p.a.mean.r | p.a.mode.r | p.b.mean.r | p.b.mode.r | CRR.r | L2.r | L3.r | L4.r | L3.mu.r | L4.mu.r | L.CV.r | median.a.d.r | mode.a.d.r | weibull_c.r | weibull_b.r | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2559 | 11 | 20 | 286.4789 | 24.66506 | 211.9293 | 168.9124 | 76.25320 | 32.78889 | 33.10929 | 32.47470 | 32.16775 | 15.72500 | 15.77696 | 15.67285 | 15.62057 | 38.00919 | 38.13158 | 37.88871 | 37.77067 | 16.04577 | 16.07842 | 16.01360 | 15.98188 | 246.6902 | 259.3793 | 259.4580 | 260.5347 | 258.7484 | 268.1751 | 20.68138 | 21.74518 | 21.75178 | 21.84204 | 21.69229 | 22.48258 | 145.2360 | 152.7066 | 152.7530 | 153.3868 | 152.3352 | 157.8850 | 31.89665 | 32.67149 | 31.15759 | 30.44960 | 12.65043 | 12.77711 | 12.52558 | 12.40361 | 38.70450 | 39.11473 | 38.33744 | 38.01243 | 12.78380 | 12.93334 | 12.63747 | 12.49658 | 14256.333 | 9777.434 | 1500.667 | 1605.642 | 0.503 | 2.4010 | 4.6700 | 7.224 | 7.856 | 8.285 | 9.248 | 10.039 | 10.827 | 11.663 | 12.072 | 12.620 | 13.126 | 13.525 | 15.02800 | 9.498822 | 10.06134 | 8.423595 | 5.564067 | 10.012 | 12.934 | 17.773 | 0.101 | 11.00303 | 3.317082 | 0.3492098 | 17.672 | 4.182 | 3.269055 | -0.8280228 | 57.05479 | 13.94600 | 95.73290 | 42.94521 | 86.01888 | 4.265055 | 0.5344524 | 143572397 | 1617370185 | 18927095193 | -2473934204 | 35199802352 | 1e-07 | 2.177178 | 3.435178 | 3.136683 | 10.61561 | 8.882625 | 10.67035 | 5.742351 | 2.067638 | 8.794915 | 0.1000084 | 19.99997 | 0.1000084 | 34.95529 | 5.912300 | 0.6656028 | 19.89997 | 10.615460 | 1.781430 | 0.1237297 | 49.45539 | 99.99993 | 50.54461 | 0 | 0.4441318 | 161478966 | 2351353463 | 36674517035 | -1951716342 | 29574870040 | 1e-07 | 5.341997 | 8.782616 | 1.532740 | 9.864254 | 14.14869 | 14.66584 | 13.60292 | 13.00865 | 13.24735 | 13.098382 | 24.85415 | 0.6629205 | 14.90152 | 3.860249 | 0.2728344 | 24.19123 | 5.473503 | 2.583600 | 0.2985818 | 42.06498 | 53.03285 | 57.93502 | 46.967080 | 0.5692686 | 305051362 | 4938483951 | 84413085236 | -8009741985 | 171321681760 | 0 | 2.696722 | 1.050308 | 4.125092 | 15.58229 |
| 2560 | 12 | 20 | 310.3521 | 24.82251 | 213.2653 | 170.4284 | 76.73561 | 31.55128 | 31.91174 | 31.19873 | 30.85627 | 15.68462 | 15.70780 | 15.66138 | 15.63810 | 37.23372 | 37.36789 | 37.10049 | 36.96879 | 16.14607 | 16.16621 | 16.12614 | 16.10643 | 278.5212 | 292.8476 | 292.9365 | 296.5834 | 294.2724 | 306.3638 | 19.92268 | 20.94746 | 20.95382 | 21.21468 | 21.04937 | 21.91428 | 152.2515 | 160.0830 | 160.1316 | 162.1252 | 160.8618 | 167.4715 | 29.35498 | 30.17865 | 28.56768 | 27.78941 | 13.58869 | 13.78073 | 13.34545 | 13.02087 | 36.13364 | 36.69115 | 35.66487 | 35.27259 | 14.04560 | 14.20998 | 13.86409 | 13.66583 | 16950.000 | 20145.792 | 1529.000 | 1611.616 | 0.571 | 2.7250 | 5.4500 | 8.167 | 8.718 | 9.215 | 10.147 | 10.844 | 11.372 | 11.939 | 12.360 | 12.803 | 13.586 | 14.332 | 15.70900 | 10.147569 | 10.66770 | 9.111729 | 6.193954 | 10.847 | 11.377 | 17.591 | 0.101 | 10.82666 | 3.290390 | 0.3242540 | 17.490 | 3.677 | 3.910328 | -1.0263343 | 59.70916 | 40.07179 | 96.22142 | 40.29084 | 59.89833 | 3.776970 | 0.5768614 | 163020972 | 1916656950 | 23302716336 | -3046140794 | 46225758098 | 1e-07 | 1.887569 | 1.229431 | 3.406567 | 11.29426 | 9.876787 | 11.45792 | 7.051412 | 2.883396 | 10.152802 | 0.1000020 | 19.99997 | 0.1000020 | 33.73304 | 5.808015 | 0.5880470 | 19.89997 | 9.963760 | 1.849015 | -0.0444131 | 51.63187 | 99.99993 | 48.36813 | 0 | 0.4938401 | 188067386 | 2799595313 | 44437689036 | -2772907266 | 43910514235 | 1e-07 | 4.806447 | 9.776785 | 1.755560 | 11.091832 | 15.13936 | 15.65515 | 14.56380 | 13.87074 | 14.68149 | 11.392439 | 25.97687 | 0.6149849 | 15.88368 | 3.985434 | 0.2632499 | 25.36188 | 6.475083 | 2.525968 | 0.0540181 | 45.86457 | 80.26023 | 54.13543 | 19.739704 | 0.5828014 | 351088358 | 6009103904 | 107754837364 | -9936643458 | 226675724189 | 0 | 3.239521 | 3.746917 | 4.291125 | 16.63549 |
| 2561 | 13 | 20 | 358.0986 | 26.99181 | 244.0854 | 196.0309 | 88.25248 | 30.65111 | 30.97916 | 30.32416 | 29.99877 | 16.56222 | 16.62685 | 16.49787 | 16.43399 | 36.28503 | 36.40635 | 36.16509 | 36.04694 | 17.45026 | 17.50284 | 17.39668 | 17.34230 | 246.6902 | 259.3793 | 259.4580 | 255.9155 | 254.3892 | 267.6363 | 15.99621 | 16.81901 | 16.82412 | 16.59441 | 16.49544 | 17.35443 | 123.7290 | 130.0934 | 130.1328 | 128.3561 | 127.5905 | 134.2347 | 28.09787 | 28.73344 | 27.44249 | 26.77323 | 13.80882 | 13.94154 | 13.67060 | 13.52743 | 34.37567 | 34.53079 | 34.22309 | 34.07339 | 14.52437 | 14.60481 | 14.44138 | 14.35609 | 13008.500 | 17940.994 | 1275.000 | 1394.355 | 0.583 | 2.5130 | 4.8500 | 7.940 | 8.712 | 9.317 | 10.587 | 11.266 | 11.796 | 12.335 | 12.651 | 12.989 | 14.029 | 14.874 | 16.40200 | 10.364617 | 10.95352 | 9.208809 | 6.160306 | 11.266 | 11.862 | 18.915 | 0.101 | 12.55426 | 3.543199 | 0.3418552 | 18.814 | 3.939 | 3.606888 | -0.9508453 | 62.09272 | 38.57363 | 96.01505 | 37.90728 | 61.40006 | 3.983775 | 0.5479576 | 163722283 | 1994322322 | 25172528270 | -3096431943 | 48018428978 | 1e-07 | 2.085617 | 1.497383 | 3.211962 | 11.57003 | 10.191565 | 11.55485 | 8.352277 | 6.224284 | 10.326048 | 0.5452360 | 19.99992 | 0.5452360 | 29.64660 | 5.444869 | 0.5342525 | 19.45468 | 9.254569 | 1.790222 | 0.0777802 | 50.49330 | 99.99993 | 49.50670 | 0 | 0.5095804 | 182191996 | 2698559726 | 42779529019 | -2871903068 | 46312819801 | 1e-07 | 4.590674 | 9.646329 | 1.952156 | 11.494005 | 15.43861 | 15.92150 | 14.90807 | 14.29910 | 14.96905 | 11.396051 | 26.89557 | 1.4374041 | 15.14360 | 3.891478 | 0.2520615 | 25.45816 | 5.717303 | 2.620821 | 0.0727989 | 46.68826 | 86.39205 | 53.31174 | 13.607874 | 0.5740204 | 345914279 | 5984357012 | 108257760931 | -10036937746 | 233390206831 | 0 | 2.851825 | 4.042555 | 4.501368 | 16.91736 |
| 2562 | 14 | 20 | 326.2676 | 26.33464 | 237.9450 | 190.6221 | 86.02561 | 31.82439 | 32.05765 | 31.58643 | 31.34362 | 16.50488 | 16.57180 | 16.43725 | 16.36921 | 36.37367 | 36.43150 | 36.31820 | 36.26505 | 17.35692 | 17.41033 | 17.30122 | 17.24326 | 294.4366 | 309.5818 | 309.6757 | 321.4368 | 317.4355 | 324.9851 | 21.44220 | 22.54513 | 22.55197 | 23.40847 | 23.11708 | 23.66687 | 164.5789 | 173.0444 | 173.0969 | 179.6709 | 177.4344 | 181.6543 | 30.00426 | 30.45045 | 29.56737 | 29.14418 | 13.59718 | 13.87527 | 12.99057 | 11.23805 | 36.11562 | 36.24826 | 35.98738 | 35.86391 | 14.12081 | 14.16125 | 14.07725 | 14.03035 | 19581.000 | 18593.551 | 1679.333 | 1730.414 | 0.664 | 2.9189 | 5.7390 | 8.515 | 9.120 | 9.706 | 10.623 | 11.250 | 11.907 | 12.563 | 12.916 | 13.336 | 14.203 | 15.009 | 16.13600 | 10.626730 | 11.15787 | 9.583030 | 6.692478 | 11.252 | 11.947 | 19.481 | 0.101 | 11.57074 | 3.401579 | 0.3200965 | 19.380 | 3.794 | 3.961588 | -1.0479797 | 59.97503 | 39.17448 | 96.56767 | 40.02497 | 60.80192 | 3.430308 | 0.5454920 | 209490136 | 2570590219 | 32592792092 | -4107992892 | 65267931931 | 1e-07 | 1.984270 | 1.320270 | 3.455819 | 11.81871 | 10.273074 | 11.72287 | 7.960387 | 4.273023 | 9.915276 | 0.1000310 | 20.00000 | 0.1000310 | 31.88965 | 5.647093 | 0.5496984 | 19.89996 | 9.347442 | 1.922395 | 0.0446746 | 47.52190 | 99.99994 | 52.47810 | 0 | 0.5136538 | 231243050 | 3491619887 | 56565658882 | -3635109042 | 59513819528 | 0e+00 | 4.563232 | 10.173043 | 1.891395 | 11.575362 | 15.68101 | 16.18406 | 15.12666 | 14.47760 | 15.28735 | 8.591582 | 26.04291 | 1.6018739 | 16.02964 | 4.003703 | 0.2553217 | 24.44104 | 5.813319 | 2.588043 | 0.0760458 | 46.19425 | 97.16024 | 53.80575 | 2.839701 | 0.6021221 | 440733186 | 7765273419 | 143169937397 | -12968148341 | 306342704434 | 0 | 2.911257 | 7.089431 | 4.438153 | 17.19710 |
| 2563 | 15 | 20 | 334.2254 | 26.67290 | 240.8075 | 192.9812 | 87.05307 | 31.64286 | 31.87649 | 31.40932 | 31.17674 | 16.51905 | 16.58523 | 16.44784 | 16.37013 | 36.27554 | 36.33279 | 36.22188 | 36.17161 | 17.47622 | 17.52059 | 17.43296 | 17.39087 | 238.7324 | 251.0123 | 251.0884 | 259.4727 | 256.5049 | 256.6323 | 16.59437 | 17.44794 | 17.45324 | 18.03603 | 17.82974 | 17.83859 | 131.9304 | 138.7166 | 138.7587 | 143.3921 | 141.7520 | 141.8224 | 29.36490 | 29.74950 | 28.99923 | 28.65520 | 14.37034 | 14.49912 | 14.23496 | 14.09320 | 34.15560 | 34.34168 | 33.97126 | 33.79001 | 14.78784 | 14.94781 | 14.61244 | 14.42169 | 9236.667 | 9075.978 | 1201.667 | 1241.396 | 0.626 | 2.7690 | 5.6178 | 8.942 | 9.568 | 10.088 | 11.015 | 11.820 | 12.512 | 13.182 | 13.440 | 13.741 | 14.614 | 15.136 | 16.25800 | 10.982258 | 11.54074 | 9.838005 | 6.621983 | 11.799 | 13.401 | 20.245 | 0.101 | 12.57862 | 3.546635 | 0.3229423 | 20.144 | 3.884 | 4.031515 | -1.1552750 | 60.19676 | 25.80000 | 96.31534 | 39.80324 | 74.17410 | 3.682199 | 0.5424677 | 184063068 | 2332018798 | 30435513757 | -3732263096 | 61191217784 | 1e-07 | 2.142742 | 2.418742 | 3.421984 | 12.22040 | 9.448153 | 11.09171 | 6.694392 | 2.933080 | 9.323490 | 0.1000019 | 20.00000 | 0.1000019 | 33.75845 | 5.810202 | 0.6149564 | 19.90000 | 10.312777 | 1.772333 | 0.0504223 | 49.34366 | 99.99993 | 50.65634 | 0 | 0.4724077 | 170018638 | 2501603522 | 39308118584 | -2317481170 | 35828894184 | 1e-07 | 5.157570 | 9.348151 | 1.671244 | 10.576315 | 15.54378 | 16.00671 | 15.01593 | 14.34900 | 15.08682 | 13.174380 | 27.73252 | 2.1417932 | 14.60546 | 3.821709 | 0.2458674 | 25.59073 | 4.942452 | 3.094322 | 0.0077833 | 44.61860 | 75.01833 | 55.38140 | 24.981602 | 0.5604893 | 354081706 | 6131851416 | 110882537246 | -10379448499 | 242930048830 | 0 | 2.434861 | 2.369403 | 4.626210 | 17.00557 |
| 2564 | 16 | 20 | 310.3521 | 23.85538 | 211.8378 | 169.8424 | 76.46164 | 30.97692 | 31.28390 | 30.67259 | 30.37392 | 16.18974 | 16.25163 | 16.12409 | 16.05316 | 36.18010 | 36.25185 | 36.10918 | 36.03926 | 16.93134 | 16.98705 | 16.87890 | 16.82953 | 318.3099 | 334.6830 | 334.7846 | 338.5946 | 336.0030 | 357.4331 | 21.02449 | 22.10594 | 22.11265 | 22.36431 | 22.19313 | 23.60860 | 162.7122 | 171.0817 | 171.1336 | 173.0812 | 171.7565 | 182.7110 | 28.29084 | 28.99965 | 27.55287 | 26.79226 | 13.70120 | 13.94374 | 13.38122 | 12.93075 | 36.21691 | 36.32707 | 36.10425 | 35.98921 | 15.02366 | 15.14870 | 14.90190 | 14.78418 | 22187.833 | 26385.098 | 1690.000 | 1884.641 | 0.632 | 2.7600 | 5.4520 | 8.768 | 9.430 | 9.909 | 10.720 | 11.258 | 11.803 | 12.294 | 12.580 | 12.916 | 13.831 | 14.564 | 16.19075 | 10.521718 | 11.03270 | 9.477378 | 6.514663 | 11.258 | 11.930 | 19.809 | 0.101 | 11.01390 | 3.318720 | 0.3154161 | 19.708 | 3.150 | 4.308202 | -1.1747411 | 62.62202 | 37.11315 | 96.37187 | 37.37798 | 62.85850 | 3.626607 | 0.5311585 | 167849455 | 2026457926 | 25217643949 | -3271733774 | 51422539428 | 1e-07 | 1.767282 | 1.408282 | 3.512850 | 11.69181 | 10.054045 | 11.67658 | 7.726990 | 5.116570 | 9.872622 | 0.4703062 | 19.99999 | 0.4703062 | 35.25883 | 5.937914 | 0.5905996 | 19.52968 | 10.917843 | 1.671939 | 0.0757414 | 48.78392 | 99.99993 | 51.21608 | 0 | 0.5027026 | 188013065 | 2889834451 | 47324710513 | -2781038923 | 45137050481 | 1e-07 | 5.452152 | 9.583739 | 1.747202 | 11.287794 | 15.47980 | 16.06434 | 14.82711 | 14.05150 | 14.96039 | 12.453883 | 27.16916 | 0.9449329 | 18.43877 | 4.294039 | 0.2773962 | 26.22423 | 6.647488 | 2.493410 | 0.0942334 | 45.76022 | 72.88986 | 54.23978 | 27.110064 | 0.5697564 | 355862521 | 6306167488 | 117543510235 | -10219870722 | 238710281311 | 0 | 3.276466 | 3.025920 | 4.050228 | 17.06615 |
Calculation of relative bias
In order to estimate the goodness of the TLS-based estimation of the
variables, the function relative.bias
computes the relative bias between the variables estimated from field
data and their respective TLS-based estimates. The relative bias is
calculated for each sample plot and each simulated plot (i.e. different
plot sizes and designs). Therefore, the input data for this function
(introduced in simulations) must be a list
of data frames containing the estimated variables (based on field and
TLS data) for all the simulated plots. Thus, a similar list to the
output of the simulations function (see above) is required,
that has the same description and format.
Optionally, the variables for which the relative bias will be
computed can be specified in a vector in
variables. Only, the names of the field
data based estimates can be introduced. If not otherwise specified, the
argument will be set to
c("N", "G", "V", "d", "dg", "d.0", "h", "h.0") by default.
Other possible variables are dgeom, dharm,
dg.0, dgeom.0, dharm.0,
hg, hgeom, hharm,
hg.0, hgeom.0 or hharm.0.
The arguments save.result and
dir.result define whether and to which
directory the output files should be saved. Two different output files
are generated. First, the data frames for each plot design (as shown
below for circular fixed areas) are saved as .csv files using the
write.csv function from the utils package.
Second, interactive line charts representing the relative biases are
saved as .html files by means of the saveWidget function in
the htmlwidgets
package. An example of these interactive line charts is provided
below.
bias <- relative.bias(simulations = Rioja.simulations,
variables = c("N", "G", "d", "dg", "d.0", "h", "h.0"),
save.result = FALSE, dir.result = NULL)
#> Computing relative bias for fixed area plots
#> (0.34 secs)
#> Computing relative bias for k-tree plots
#> (0.08 secs)
#> Computing relative bias for angle-count plots
#> (0.06 secs)The function calculates the relative bias between the field data
estimates (specified in variables) and the counterpart
variables that are estimated based on TLS data. The TLS counterparts for
the density (N) are the variables N.tls,
N.hn, N.hr, N.hn.cov,
N.hr.cov and N.sh for circular fixed area and
k-tree plots, and N.tls and N.pam for
angle-count plots. The same pattern applies to the basal area
(G) and the volume (V) where the corresponding
TLS-based estimates are G.tls, G.hn,
G.hr, G.hn.cov, G.hr.cov,
G.sh and G.pam, and V.tls,
V.hn, V.hr, V.hn.cov,
V.hr.cov, V.sh and V.pam
respectively. In case of mean and dominant diameters (d,
dg, dgeom, dharm,
d.0, dg.0, dgeom.0, and
dharm.0) and heights (h, hg,
hgeom, hharm, h.0,
hg.0, hgeom.0 and hharm.0), for
all three plot designs their respective counterpart variables are
d.tls, dg.tls, dgeom.tls,
dharm.tls, d.0.tls, dg.0.tls,
dgeom.0.tls and dharm.0.tls (for the
diameter), and h.tls, hg.tls,
hgeom.tls, hharm.tls, h.0.tls,
hg.0.tls, hgeom.0.tls,
hharm.0.tls and in addition P99 (for the
height). The relative bias are calculated as follows
where
is the value of the field estimate and
the value of its TLS counterpart corresponding to plot
of
sample plots. For each plot size defined by the radius, k or BAF, the
biases are calculated and stored as a data frame (shown below). Each row
represents the a simulated plot of a certain size (here defined by
radius) and the columns contain the calculate bias between
the variables indicated in the column names. The two compared variables
are joint with . as separation, e.g. N.N.tls
means that the bias between N and N.tls was
calculated.
head(bias$fixed.area)| radius | N.N.tls | N.N.hn | N.N.hr | N.N.hn.cov | N.N.hr.cov | N.N.sh | G.G.tls | G.G.hn | G.G.hr | G.G.hn.cov | G.G.hr.cov | G.G.sh | d.d.tls | dg.dg.tls | d.0.d.0.tls | h.h.tls | h.P99 | h.0.h.0.tls | h.0.P99 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2.5 | 0.00000 | 5.143769 | 5.175673 | 7.687337 | 6.637258 | 0.0000000 | -9.6703456 | -5.023997 | -4.995178 | -2.577931 | -3.554964 | -9.6703456 | -5.1950393 | -5.1950393 | -5.1950393 | -17.15280 | -9.404255 | -17.15280 | -9.404255 |
| 2.6 | 0.00000 | 5.143769 | 5.175673 | 7.687337 | 6.637258 | 0.0000000 | -9.6703456 | -5.023997 | -4.995178 | -2.577931 | -3.554964 | -9.6703456 | -5.1950393 | -5.1950393 | -5.1950393 | -17.15280 | -9.462006 | -17.15280 | -9.462006 |
| 2.7 | 50.00000 | 57.715653 | 57.763509 | 61.202593 | 59.709142 | 50.0255646 | 36.9178682 | 43.960606 | 44.004289 | 47.235110 | 45.855820 | 36.9247528 | -6.7531804 | -6.7288477 | -5.1950393 | -15.63418 | -9.513678 | -17.15280 | -9.513678 |
| 2.8 | 33.33333 | 40.191691 | 40.234230 | 43.781747 | 42.351883 | 33.4575777 | 33.2182993 | 40.070740 | 40.113242 | 43.792068 | 42.337512 | 33.3234726 | -0.8541952 | -0.8376348 | 0.2062484 | -14.02461 | -11.170292 | -15.06550 | -11.170292 |
| 2.9 | 25.00000 | 31.429711 | 31.469591 | 34.807426 | 33.467629 | 25.2969253 | 25.9246555 | 32.401928 | 32.442104 | 35.906474 | 34.537062 | 26.2018841 | -0.2929137 | -0.2804175 | 0.5072745 | -14.43016 | -14.105740 | -15.18488 | -14.105740 |
| 3.0 | 0.00000 | 5.143769 | 5.175673 | 7.378223 | 6.372637 | 0.3139353 | 0.5167802 | 5.687131 | 5.719200 | 8.226848 | 7.170623 | 0.8472341 | -3.6238902 | -3.6289551 | -3.6911015 | -14.53109 | -13.990006 | -15.31393 | -14.150873 |
For better visualization, line charts are created that show the
relative bias of a given variable and plot design. As an example, the
interactive graphic showing the relative bias of basal area estimations
(G) for fixed are plots can be seen when following the link
(RB.G.fixed.area.html).
Functions facilitating model-based or model-assisted sampling approaches
To facilitate the application of model-based sampling, two additional
functions are included in the FORTLS package. The function
correlations computes the correlations
between variables estimated from field data and those estimated from TLS
data and calculates the respective Pearson and Spearman correlation
coefficients. The results are saved as .csv files and represented as
line charts and heat maps (when applying the function
optimize.plot.design).
Computing correlations
The correlations function computes the
correlations for all the plot designs that are introduces as elements of
a list in simulations. The format and
description must be the same as the output list of the
simulations function. Also similar to the function
relative.bias, the variables for which the correlations are
to be calculated can be specified in
variables. By default, this argument is
set to
variables = c("N", "G", "V", "d", "dg", "d.0", "h", "h.0").
If only one of the two above-mentioned correlation measures should be
calculated, it can be specified in method.
This argument is set to method = c("pearson", "spearman")
by default and both correlation coefficients are computed.
fixed.area.simulations <- list(fixed.area = Rioja.simulations$fixed.area[Rioja.simulations$fixed.area$radius < 7.5, ])
cor <- correlations(simulations = fixed.area.simulations,
variables = c("N", "G", "d", "dg", "d.0", "h", "h.0"),
method = c("pearson", "spearman"),
save.result = FALSE, dir.result = NULL)
#> Computing correlations for fixed area plots
#> (33.46 secs)In addition to the calculation of the correlation measures, the
function also performs tests of association and returns the p-values.
The output of this function is a list containing the following three
elements correlations, correlations.pval and
opt.correlations. Each of them is a list including, if not
otherwise specified (in method), two elements
pearson and spearman. These two elements are
lists again that include separate data frames for each plot design
(circular fixed area, k-tree and angle-count plots). The data frames
contain the corresponding correlation coefficients (in
correlations), the calculated p-values (in
correlations.pval) and the optimal correlations for a given
plot size and field data estimate (in
opt.correlations).
cor$correlations$pearson$fixed.area[20:26,1:15]| radius | N.N.tls | N.N.hn | N.N.hr | N.N.hn.cov | N.N.hr.cov | N.N.sh | N.n.pts | N.n.pts.est | N.n.pts.red | N.n.pts.red.est | N.G.tls | N.G.hn | N.G.hr | N.G.hn.cov |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4.7 | 0.8427010 | 0.8427010 | 0.8427010 | 0.8372458 | 0.8383825 | 0.8416905 | 0.3827329 | 0.6163229 | 0.8403905 | 0.8064847 | 0.6918248 | 0.6918248 | 0.6918248 | 0.6733457 |
| 4.8 | 0.5341718 | 0.5341718 | 0.5341718 | 0.5474591 | 0.5450412 | 0.5385519 | 0.3803623 | 0.5561943 | 0.6830626 | 0.4886011 | 0.4470624 | 0.4470624 | 0.4470624 | 0.4507763 |
| 4.9 | 0.5341718 | 0.5341718 | 0.5341718 | 0.5474591 | 0.5450412 | 0.5385629 | 0.3803623 | 0.5561943 | 0.6830626 | 0.4886011 | 0.4470624 | 0.4470624 | 0.4470624 | 0.4507763 |
| 5.0 | 0.5341718 | 0.5341718 | 0.5341718 | 0.5474591 | 0.5450412 | 0.5384909 | 0.3803623 | 0.5561943 | 0.6830626 | 0.4886011 | 0.4470624 | 0.4470624 | 0.4470624 | 0.4507763 |
| 5.1 | 0.4267459 | 0.4267459 | 0.4267459 | 0.4406877 | 0.4382988 | 0.4300013 | 0.2931150 | 0.4148178 | 0.6415550 | 0.3953048 | 0.3313156 | 0.3313156 | 0.3313156 | 0.3375288 |
| 5.2 | 0.6374909 | 0.6374909 | 0.6374909 | 0.6407699 | 0.6404134 | 0.6395937 | 0.3792903 | 0.4078300 | 0.6505892 | 0.4761103 | 0.5101267 | 0.5101267 | 0.5101267 | 0.5014544 |
| 5.3 | 0.6064784 | 0.6064784 | 0.6064784 | 0.6012499 | 0.6020179 | 0.6052299 | 0.2815443 | 0.2962698 | 0.5317175 | 0.4570482 | 0.4429577 | 0.4429577 | 0.4429577 | 0.4322508 |
All mentioned data frames are divided into rows each of which
represent a given plot size defined by radius (for circular
fixed area plots), k (for k-tree plots) or BAF
(for angle-count plots). The columns of the data frames in
correlations (shown above) and
correlations.pval (shown below) contain the calculated
coefficients and p-values respectively for the corresponding
correlation. The column names are composed of the two variables
(e.g. N and N.tls) separated by .
(giving N.N.tls) that were correlated as described for the
relative.bias function.
cor$correlations.pval$pearson$fixed.area[20:26,1:15]| radius | N.N.tls | N.N.hn | N.N.hr | N.N.hn.cov | N.N.hr.cov | N.N.sh | N.n.pts | N.n.pts.est | N.n.pts.red | N.n.pts.red.est | N.G.tls | N.G.hn | N.G.hr | N.G.hn.cov |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4.7 | 0.0022053 | 0.0022053 | 0.0022053 | 0.0025099 | 0.0024441 | 0.0022596 | 0.2750097 | 0.0577525 | 0.0023308 | 0.0048223 | 0.0266593 | 0.0266593 | 0.0266593 | 0.0328198 |
| 4.8 | 0.0905190 | 0.0905190 | 0.0905190 | 0.0813023 | 0.0829309 | 0.0874083 | 0.2485167 | 0.0755965 | 0.0205199 | 0.1272621 | 0.1680119 | 0.1680119 | 0.1680119 | 0.1640786 |
| 4.9 | 0.0905190 | 0.0905190 | 0.0905190 | 0.0813023 | 0.0829309 | 0.0874006 | 0.2485167 | 0.0755965 | 0.0205199 | 0.1272621 | 0.1680119 | 0.1680119 | 0.1680119 | 0.1640786 |
| 5.0 | 0.0905190 | 0.0905190 | 0.0905190 | 0.0813023 | 0.0829309 | 0.0874511 | 0.2485167 | 0.0755965 | 0.0205199 | 0.1272621 | 0.1680119 | 0.1680119 | 0.1680119 | 0.1640786 |
| 5.1 | 0.1905500 | 0.1905500 | 0.1905500 | 0.1748975 | 0.1775217 | 0.1868221 | 0.3816974 | 0.2045907 | 0.0333605 | 0.2288530 | 0.3195963 | 0.3195963 | 0.3195963 | 0.3100452 |
| 5.2 | 0.0348622 | 0.0348622 | 0.0348622 | 0.0336470 | 0.0337777 | 0.0340795 | 0.2499634 | 0.2130948 | 0.0301856 | 0.1387719 | 0.1088955 | 0.1088955 | 0.1088955 | 0.1160739 |
| 5.3 | 0.0479069 | 0.0479069 | 0.0479069 | 0.0503955 | 0.0500245 | 0.0484933 | 0.4016223 | 0.3763530 | 0.0922936 | 0.1575664 | 0.1724261 | 0.1724261 | 0.1724261 | 0.1842722 |
The data frames of the opt.correlations list (example
shown below) are also divided into rows that represent the different
plot sizes. For a given plot size and variable (specified in the
argument variables), the best correlating TLS-based
estimate and the corresponding correlation coefficient is displayed in
this table. The columns named <variable>.metric (with
<variable> being here N, G,
d, dg, d.0, h and
h.0) contain the TLS-based variable or metric that yielded
the best correlation with the respective field data-based variable of
the column name for a certain plot radius. The columns
<variable>.cor display the measures of the respective
correlations. That means, in the example shown here, the TLS-based
estimate that yielded the best correlation with the field data based
variable density (N) for circular fixed area plots with a
radius of 4.4 m is ID.rho. And the correlation coefficient
is 0.7286.
cor$opt.correlations$pearson$fixed.area[20:26,]| radius | N.cor | N.metric | G.cor | G.metric | d.cor | d.metric | dg.cor | dg.metric | d.0.cor | d.0.metric | h.cor | h.metric | h.0.cor | h.0.metric | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 20 | 4.7 | 0.8427010 | N.hn | 0.8673762 | G.hr.cov | 0.8661827 | d.tls | 0.8480509 | dg.tls | 0.8243517 | d.0.tls | 0.9542529 | h.tls | 0.8614859 | hg.tls |
| 21 | 4.8 | 0.7697247 | p.b.mode.r | 0.7355321 | V.hn.cov | 0.7636362 | dg.tls | 0.7533423 | dg.tls | 0.8279615 | d.0.tls | 0.8425797 | P95 | 0.8115912 | h.0.tls |
| 22 | 4.9 | -0.7663825 | p.a.mode.r | 0.7355321 | V.hn.cov | 0.7636362 | dg.tls | 0.7533423 | dg.tls | 0.8279615 | d.0.tls | 0.8443094 | P95 | 0.8115912 | h.0.tls |
| 23 | 5.0 | -0.7626689 | p.a.mode.r | 0.7355321 | V.hn.cov | 0.7636362 | dg.tls | 0.7533423 | dg.tls | 0.8279615 | d.0.tls | 0.8449040 | P95 | 0.8115912 | h.0.tls |
| 24 | 5.1 | -0.7979226 | p.a.mode.r | -0.6614491 | p.a.mode.r | 0.7653580 | dg.tls | 0.7572720 | dg.tls | 0.8279615 | d.0.tls | 0.8572419 | P95 | 0.8115912 | h.0.tls |
| 25 | 5.2 | -0.7586088 | mean.h.z | -0.6689808 | P05 | 0.7948788 | dg.tls | 0.7840312 | dg.tls | 0.8279615 | d.0.tls | 0.9003149 | hg.tls | 0.8115912 | h.0.tls |
| 26 | 5.3 | -0.7823391 | mean.h.z | -0.7073419 | P05 | 0.8182592 | dg.tls | 0.8155215 | dg.tls | 0.8279615 | d.0.tls | 0.8918826 | hg.tls | 0.8115912 | h.0.tls |
The correlations functions creates different files and
saves them (if save.result = TRUE, default setting) to the
directory indicated in dir.result. These files are, on the
one hand, .csv files of the data frames in the lists
correlations and opt.correlations created by
means of the write.csv function from the utils package.
These .csv files will be named as
correlations.<plot design>.<method>.csv and
opt.correlations.<plot design>.plot.<method>.csv
with <plot design> being
fixed.area.plot, k.tree.plot or
angle.count.plot and <method> being
pearson or spearman. On the other hand,
interactive line charts representing the correlation coefficients will
be created for each variable (selected in variables) as
.html file using the saveWidget function in the htmlwidgets
package. As an example, the interactive line chart for the variable
height (h) and fixed area plots (pearson measure) is shown
(correlations.h.fixed.area.pearson.html).
Visualizing correlations
In order to visualize the optimal correlations, the function
optimize.plot.design creates heat maps and
is applied as follows:
optimize.plot.design(correlations = cor$opt.correlations,
variables = c("N", "G", "d", "dg", "d.0", "h", "h.0"),
dir.result = NULL)The function creates the heat maps based on the optimal correlation
list (opt.correlations from the output of the
correlations function) introduced in
correlations. The introduced list must
have the same format and description as the opt.correlation
list. Similar to the other functions described above, the variables of
interest can be selected in variables
(default setting:
variables = c("N", "G", "V", "d", "dg", "d.0", "h", "h.0")).
This function generates interactive heat maps with the
saveWidget function of the htmlwidgets
package and saves these graphics to the directory indicated in
dir.result (or by default to the working
directory). For each plot design and correlation measure a plot is
generated and named as
opt.correlations.<plot design>.<method>.html
where <plot design> can be
fixed.area.plot, k.tree.plot or
angle.count.plot and <method> either
pearson or spearman according to the plot
design and correlation measure.
As an example, the heat map of the pearson correlation coefficient for fixed area plots and pearson measure is provided and can be seen when opening the link (opt.correlations.fixed.area.pearson.html).